Experience Chromogenic Differentiation

Quick Nav
Categories
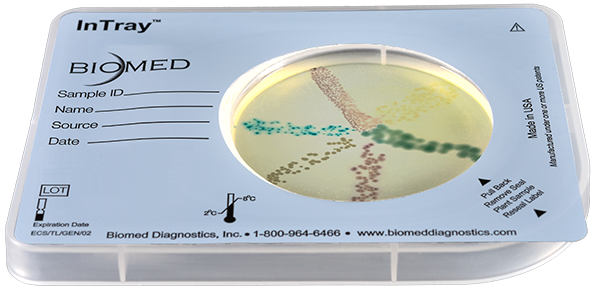
InTray® COLOREX™ Screen
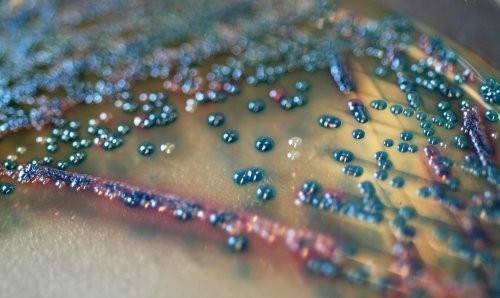
For Rapid Isolation and Differentiation of Urinary Tract Pathogens
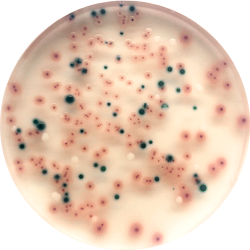
The major target of this medium is the detection of urinary tract pathogens, but COLOREX Screen has a broader application as a general nutrient agar for the isolation of various microorganisms.
COLOREX Screen can also be used to differentiate various microorganisms in other infected areas; e.g., scars, wounds, etc. In addition, COLOREX Screen is useful when supplemented with various antibiotics in detecting increasingly important nosocomial and multidrug resistant microorganisms.
For chromogenic InTrays with antibiotic supplementals, please see:
Specifications
Step-by-Step
More Information
VTM-C19 transport media is designed for specimen preservation in preparation for analysis with validated qRT-PCR assays to detect severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) that causes COVID-19 disease in humans.
This is CDC-approved media for COVID-19 sample collection and transport. VTM-C19 has been validated for stable transport of SARS-CoV-2 only.
Biomed Diagnostics also offers Saline .85% transport media for a general viral transport – our R&D lab has validated the Saline .85% for SARS-CoV-2, and the Saline is checked throughout the shelf life of the product for sterility by our QC Labs
Room Temperature validation is ongoing and will be update monthly.
Practical Bench Comparison of BBL CHROMagar Orientation and Standard Two-Plate Media for Urine Culture
John Merlino, Steven Siarakas, Graham Robertson, Glenn Funnell, Thomas Gottlieb, and Ross Bradbury
J Clin Microbiol; 34.
Read Paper ►
Evaluation of Use of a New Chromogenic Agar in Detection of Urinary Tract Pathogens
Z. Samra,M. Heifetz, J. Talmore, E. Bain, and J. Bahar
JOCM Vol 36, No 4
Read Paper ►
Evaluation of CHROMagar Orientation for Differentiation and Presumptive Identification of Gram-Negative Bacilli and Enterococcus Species
Z. Samra,M. Heifetz, J. Talmore, E. Bain, and J. Bahar
J Clin Microbiol; 34.
Read Paper ►
A Study on Evaluation of Different Culture Media for the Isolation of Routine Urinary Pathogens
Biji, S. Manjusree, O. Sasikumari and J.T. Ramani Bai/h6>
IJCM Vol 6 No 2
Read Paper ►
The assay results demonstrate consistent amplification and Cq quantification of the nCov-2 N gene (using the N1 CDC primer) in the solution after incubations. In addition, excellent dilution linearity is demonstrated across all samples and replicates (See Figure 1) indicating consistency in the performance of the solution Saline Solution (0.85%) replicate samples and incubation time. This data indicates that the Biomed ISO 13845 manufactured Saline Solution (0.85%) does not negatively interfere with the qRT-PCR detection of Sars-Cov-2 viral nucleic acid materials after incubation at room temperature 25°C for 72 hours. A delay in amplification signal was observed after 168-hour (7 days) incubation at 25°C, probably due to specimen degradation. However, incubation at 4°C for 168 hours (7 days) did not negatively interfere with the qRT-PCR performance. In conclusion, the result indicates that the Saline Solution (0.85%) is compatible with the designated viral specimen inoculation, nucleic acid extraction and qRT–PCR assay when used as described herein.
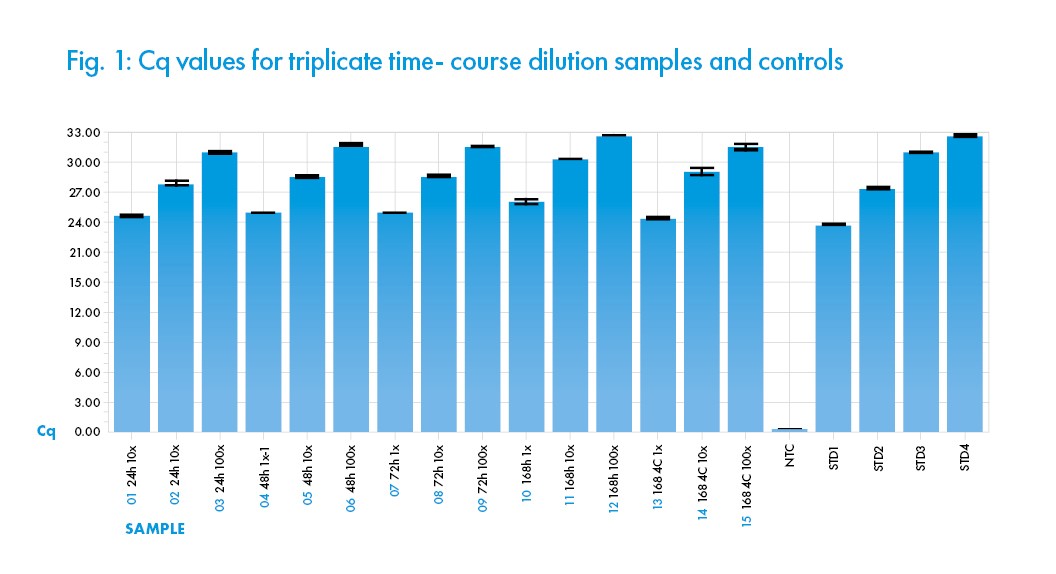
All samples shown are means from triplicate assays, with error bars indicating the arithmetic error for Cq shown. The performance of the qRT-PCR reaction assessed with standard curves indicated R2 of 0.98, Efficiency of 114%, and Slope of -3.033. All the values fall within acceptable quantitative PCR ranges.